Genomics Sandbox¶
In this app you will find material for the Genomics sandbox of the Health Data Science sandbox. It contains course tutorials, datasets, and tools you can use for research or self-learning. Each course item of this sandbox is based on Jupyterlab. Jupyterlab is a web-based integrated development environment for Jupyter notebooks, code, and data.
Available material¶
Items are periodically added to this app and can be chosen from the menu. Each item can a be course, a setup to work with specific software, a research example, and comes with all necessary packages already installed, notebooks with code and explanations, and a dedicated webpage with additional material (notes, slides, recordings, ...).
Courses¶
The available courses are:
Course Name |
Description |
Link |
Programming Language |
|---|---|---|---|
Introduction to NGS |
A one-week course to introduce NGS data, from data alignment to bioinformatics analysis |
Python, R, Bash |
|
Introduction to |
A course introducing and applying bioinformatic tools to perform a whole population genomics analysis |
Python, R, Bash |
|
Introduction to GWAS |
An introductory course in Genome-Wide Association Studies |
R, Bash |
Choosing an Item¶
You can choose a course or a tool from the drop down menu accessible in the job submission page (red circle in the figure below).
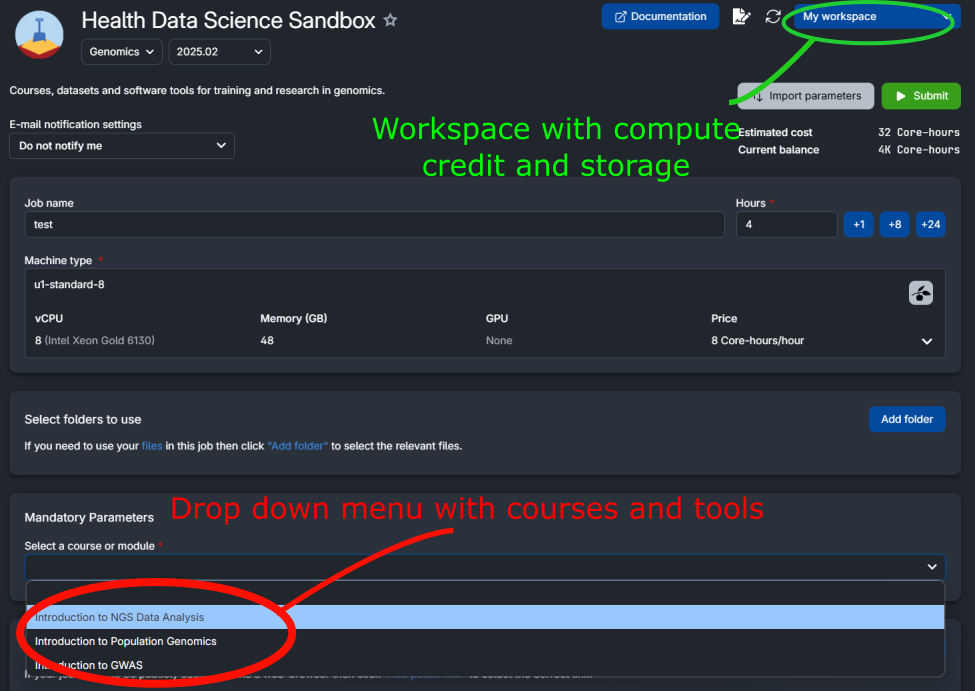
Note
The App needs to download data and packages. Depending on the selected course/tool, this can take some time. See below how to reuse data and avoid long waiting times (you still need to download data the first time you run the app).
Available options¶
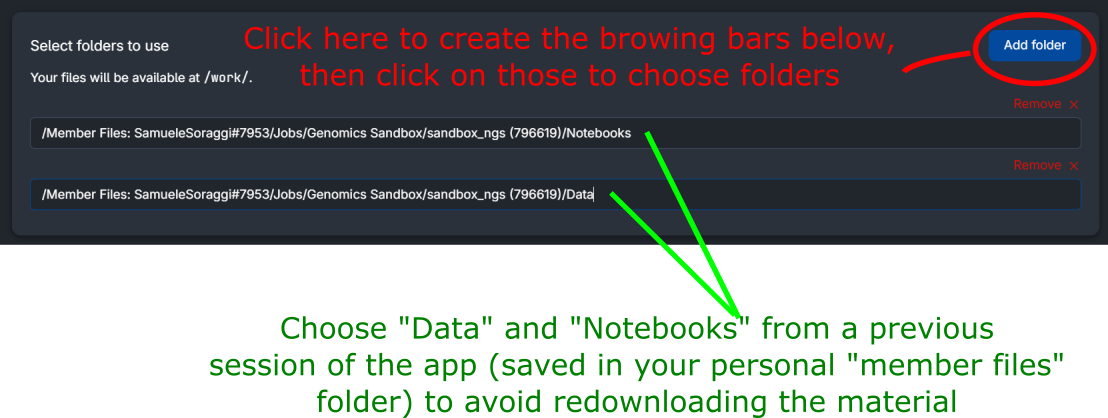
You can use data and notebooks running in a previous session of the app, otherwise, all the datasets the notebooks will be downloaded every time a new session is started.
To choose the data from previous sessions, click on Add folder (red circle in the figure below). Then, you need to select your latest session of the sandbox (inside the folder Jobs/Genomics Sandbox, as shown in the example below) and choose the folder you need. Accepted folders are Data and Notebooks (such as the two folders chosen in the figure below).
Download Course Data and Notebooks¶
Each course is open-source and the datasets are freely available. Here you have the link to all repositories to download the datasets.
Course Name |
Data Repository |
|---|---|
Introduction to NGS Data Analysis |
|
Introduction to Population Genomics |
|
Introduction to GWAS |
Download Generated Data¶
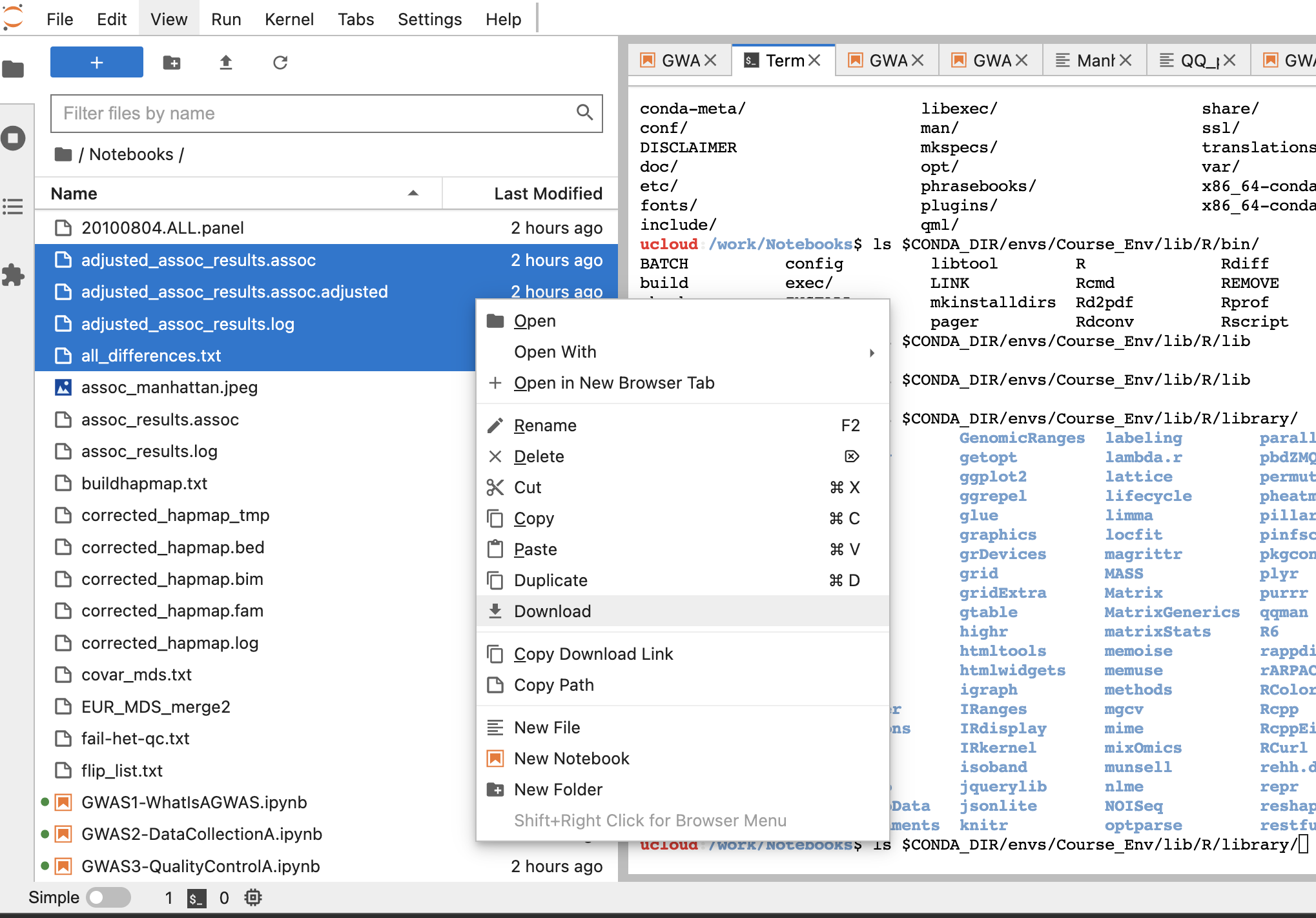
You can easily download the generated files by right-clicking the selected files in JupyterLab and choosing download (see figure below).
Contents